|
|
CURRENT RESEARCH LINES
-
CHARACTERIZATION OF THE METABOLIC AND FUNCTIONAL ACTIVITY OF LACTIC BACTERIA
- Characterization of enzymatic activities of probiotic bacteria associated to their functionality.
|
Gut microbiota can provide their host some metabolic capacity lacking in the cells of the human gut, such
as the fermentation of non-digestible dietary compounds, methanogenesis, gluconeogenesis, xenobiotic degradation, and biosynthesis of essential
amino acids, vitamins, and isoprenoids. The fermentation of non-digestible dietary compounds - prebiotics- by intestinal microbiota allows the
recovery and storage of energy as short-chain fatty acids, which leads to a decrease in pH, facilitates the absorption of ions, and provides
epithelial cells with a source of direct energy. Furthermore, the microbiota in the colon can degrade non-digestible dietary components releasing
their bioactive elements that only this way can be absorbed and exert their effect.
In collaboration with other groups from the institute, we have studied the capacity Lactobacillus,
Streptococcus, and Bifidobacterium strains have to use lactulose and its pure trisaccharide derivatives, as well as protein glycoconjugates.
It was observed that these compounds can be used when found as the sole carbon source. The capacity of the probiotic bacteria L. plantarum, L.
casei, L. acidophilus, and B. lactis to degrade certain plant-derived polyphenols, such as anthocyanins, due to the presence of β-glycosidase
activity was also showed. Similarly, wine polyphenols and particularly oligomers with mean degree of polymerization and polymeric flavonoids
(procyanidins), can modulate gut microbiota or be metabolized by it, as in the case of L. plantarum IFPL 935 from our collection, which shows
an activity only possible by few intestinal species.
|
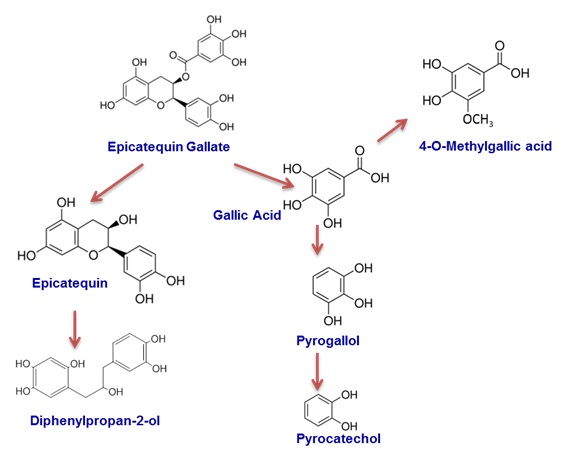
Degradation of flavan-3-ol by Lactobacillus plantarum IFPL935 |
-
Assessment of food ingredients (probiotic and prebiotics) potentially associated to intestinal balance
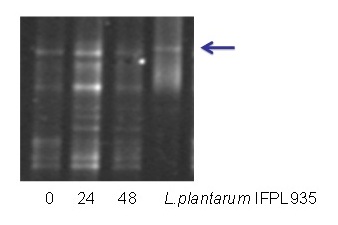
PCR-DGGE detection of Lactobacillus plantarum IFPL935 during its incubation with colonic
microbiota |
Due to the special interest of some L. plantarum strains, e.g., the previously mentioned IFPL935, the
Group is engaged in studying the role this strain has in the colonic metabolism of potentially bioactive compounds by adding this lactobacillus
to complex cultures as well as a polyphenol-enriched red wine extract. Changes in the main colonic microbiota groups and the metabolic activity
(short-chain fatty acids, ammonia, and polyphenol catabolism) in presence of IFPL935 were studied.
|
-
Colonic microbiota in conditions of normobiosis and obesity-associated dysbiosis
|
The dysbiosis in obese individuals characterizes by a lower diversity of intestinal microbiota, in terms
of microbial groups as well as genetic functionality. This has been related to an increased intestinal permeability caused by dietary fat,
as well as to certain inflammatory and hormonal responses of the intestinal epithelium. There are limitations for studying the mechanism of
interaction between the microbiota and the human intestinal mucosa due to the difficult physiological access to the different areas in the gut
where these interactions take place.
In this line of research, the Group focuses on the study of intestinal microbiota-host interactions.
For this, we develop in vitro gastro intestinal simulators that reproduce the physiological conditions of the intestinal tract. We study
the possible links microbial dysbiosis, metabolic disorders and cognition.
|

Dynamic gastro intestinal simulator |
-
Molecular tools for the detection, characterization, identification, and expression of functional
features of interest in lactic bacteria
Over the years, the Group has created a collection of lactic bacteria made up by approximately 500 strains
corresponding to the Lactococcus, Lactobacillus, Leuconostoc, and Enterococcus genera, most of which have been isolated from dairy products
produced by artisanal fermentation. An important part of these strains have been identified through molecular methods, mainly 16S- rDNA PCR
and quantitative PCR, and have additionally been characterized at molecular level based on their proteolytic system, amino acid catabolism,
aroma development, and bacteriocin production.
Currently, one of the goals of the Group in this line of research is to have a reliable system to complete
the identification of the collection of CIAL´s microorganisms in a rapid way and with a wide processing capacity. Fort this, we have optimized
the identification using the MALDI-TOF/TOF mass spectrometry, which identifies the strains considering their peptide profile characterized by
molecular mass (m), charge (z), and mass-to-charge ratio (m/z). Additionally, previous studies on antimicrobial activity characterization has
allowed us to broaden the usefulness of the MALDI-TOF technique to classify the strains based on bacteriocin production. The technique has
allowed rapidly differentiating lacticin 3147- and nisin-producing strains.
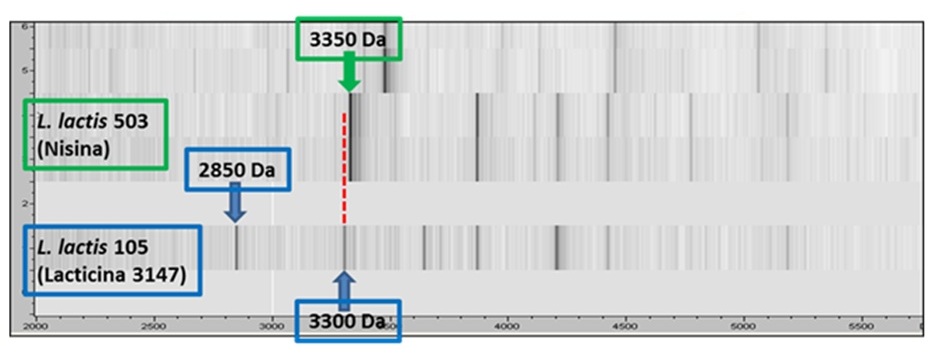
Identification of bacteriocin-producing lactic acid bacterial strains
In collaboration with the Biological Research Centre (CIB, CSIC), the Group has developed and registered
transcriptional fusion vectors that allow detecting uni- and bi-directional promoter regions in lactic bacteria. The vectors contain as a marker
a synthetic mRFP gene that codifies the mCherry red fluorescent protein (pTLR) and the GFP gene that codifies the green fluorescent GFP (pTLGR)
protein. Plasmids are of great use for characterizing promoter regions and for the expression of divergent genes. These vectors allow studying the
environmental conditions involved in genetic regulation in real time, as well as the interaction mechanisms between bacteria.
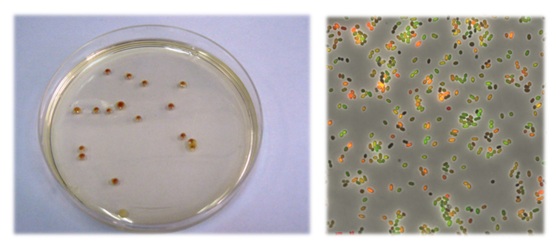
Emission of red and green fluorescence during the growth of bacteria with the transcriptional
fusion pTLGR vector.
CONSOLIDATED RESEARCH LINES IN WHICH THE GROUPS HAS EXPERIENCE
-
Characterization of lactic bacteria metabolism in relation with their technological ability
Since the creation of the Group, diverse studies on the characterization of the proteolytic system
of lactic bacteria and their use in fermented dairy products have been carried out. After gaining experience in the management of enzymatic
techniques, we specialized in the study of lactic bacteria metabolism, identification of genes that codify for enzymatic activities of interest,
as well as the characterization of these activities, with the aim of applying them in the improvement of technological processes, such as
cheese ripening.
The studies on amino acid catabolism and flavour production by lactic bacteria began with the granting
of an European project in which the members of the consortium were pioneers in this topic in Europe. Several national projects followed this
first one, aimed at enhancing the aroma through the induction by lytic agents. One of these agents, the bacteriocin lacticin 3147, produced
by a lactococcus from our collection, exerts lytic activity on sensitive bacteria and was widely studied. Studies led to the complete molecular
characterization, to understand its mechanism of action, and the construction of a transconjugant carrying the plasmid that included the genes
of the bacteriocin protected by a patent. This lactococcus can be used as a culture starter to speed up the ripeningof cheese and as a bioprotective
culture to prevent late blowing in semi-hard cheeses by Clostridium.
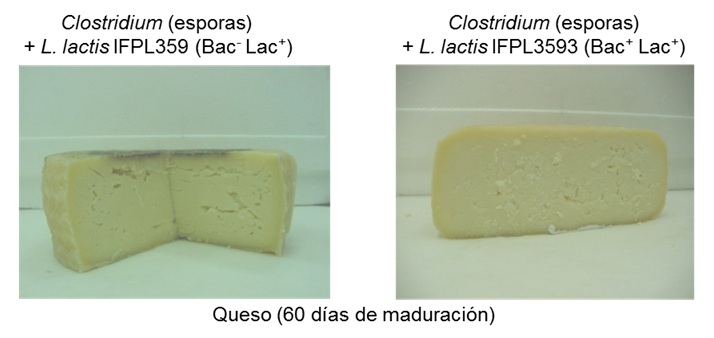
Prevention of delayed swelling in cheese using L. lactis producer lacticin 3147
|




|















